Researchers today collectively generate a massive amount of digital information during the course of their daily activities, but a lack of workflows for capturing and sharing metadata (data about data) can result in unnecessary information loss. A growing number of scientists and funding organizations have called for “FAIR” (findable, accessible, interoperable, reusable) data to extend the shelf life of research output. The FAIR Guiding Principles describe how to use metadata and web infrastructure to preserve and facilitate the exchange of scientific data.
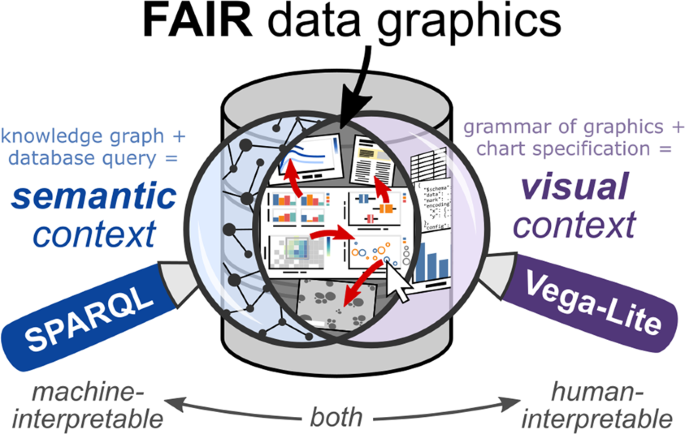
Improving the machine-interpretability of data does not need to come at the cost of human-interpretability. In our recent paper titled “FAIR and Interactive Data Graphics from a Scientific Knowledge Graph,” we describe how the fusion of readily available technologies from the Semantic Web and data visualization communities provides a powerful platform for interactive, visual exploration of linked scientific data. The approach enables the interactive exploration of data and metadata in a way not possible with conventional plotting tools.
A knowledge graph is a type of database that uses a graph, or network, to describe the relationships between objects, with the goal of representing domain knowledge. Examples of knowledge graphs include DBpedia, a linked collection of cleaned data from Wikipedia, and MaterialsMine, a curated database of polymer nanocomposite materials data used as the main data source for the demonstrations in our article. These databases can be searched using SPARQL, the graph-based query language for the Semantic Web. SPARQL provides a concise way to retrieve data from a knowledge graph, and by examining a SPARQL query, one can better understand the meaning, or semantics, of the data returned by the query.
To enable exploratory visualization of a knowledge graph, we turned to Vega-Lite, a grammar for interactive data graphics developed by the University of Washington Interactive Data Lab. Building upon Leland Wilkinson’s grammar of graphics—the paradigm behind declarative plotting tools such as ggplot2 and Observable Plot—Vega-Lite uses a compact JSON format to specify a chart from a dataset, including modes of interactivity such as tooltips and selections. Altair, a project built on top of Vega-Lite, supports the generation of Vega-Lite chart specifications using the Python programming language. Instead of considering a chart as one of a set of predefined types (bar chart, pie chart, scatter plot, line chart, etc.), the grammar of graphics illuminates the deeper structure of data visualizations, wherein a data mark (point, line, text, etc.) has one or more of its visual properties (color, size, x and y position, etc.) defined through encoding channels driven by the mapping of data attributes along discrete or continuous scales. Compared to many existing plotting tools today, this philosophy represents an unconventional but powerful way to describe the transformation of raw data into visual representations. Additionally, the smart default behavior programmed into Vega-Lite allows for the specification of a chart with minimal syntax while providing the freedom to override these defaults and customize the chart specification incrementally as needed.
Since SPARQL and Vega-Lite are web-based technologies, we found that the best example of their complementarity came from the use of HTTP URIs (uniform resource identifiers, such as a URL) as data attributes encoded through hyperlink (“href”) or image URL (“url”) encoding channels. URIs of items in the knowledge graph can be returned by a SPARQL query and plotted as data marks in Vega-Lite so that when the interactive chart is rendered, someone viewing the chart can click on a data mark to visit the HTML page for that resource. Moreover, we model chart objects themselves as resources in the knowledge graph, each with a URI able to be queried using SPARQL and further plotted in Vega-Lite inside a “meta-chart” (chart about charts). As more scientific data become available with URIs as identifiers, this approach lends itself well to making data graphics interoperable (the “I” in FAIR) from the perspective of exploratory and explanatory data visualization.
As a demonstration of interoperability, we show a chart with a federated SPARQL query that joins information from MaterialsMine and DBpedia and plots the Vega-Lite chart in Observable, a computational notebook platform that runs in a web browser. Computational notebooks such as Jupyter, R Markdown, and Observable notebooks play an important role in modern scientific communication by allowing the near seamless integration of prose, code, and data. Jeffrey Perkel, technology editor at Springer Nature, has written about the benefits of computational notebooks for scientific writing, as well as the promising qualities of reactive notebook platforms such as Observable. These notebooks have the potential to streamline scientific research and communication, even for those who would not consider themselves “programmers.”
While we believe SPARQL and Vega-Lite represent best-in-class tools for exploratory visualization of data in a knowledge graph today, we also discuss in our article the practical challenges around portability, persistence, and performance. Interactive, web-based data visualizations require an internet-connected device and a modern web browser, whereas much scientific communication today happens in the form of static documents including manuscripts, presentation slides, and posters. In addition, issues around version control of vocabularies and ontologies could impact the persistence of metadata-defined charts. For example, changes to the way data are modeled might impact the way those data can be queried. Lastly, the development and maintenance of a robust and performant database with a public-facing endpoint is non-trivial and requires support to keep data in FAIR condition. Barend Mons writes that 5% of all research funds should go towards data stewardship, a bold but necessary claim in our modern climate of data-intensive scientific research. Funding mandates aside, continued embracement of the FAIR principles requires a transformation in data culture, including the expectations set by scientists concerning the generation, consumption, and sharing of data.
Most conversations around FAIR guiding principles and data stewardship revolve around the machine-interpretability of data. While the exchange of scientific findings involves a plethora of digital tools and infrastructure, scientific communication remains at its heart a human endeavor. Curators of data and creators of data visualizations still bear the responsibility to present data clearly and accurately—this task can be nontrivial, as discussed in a recent news feature in Nature—but we hope that an infrastructure with greater transparency into the semantic and visual context of data increases confidence and trust in scientific data. In our article, we propose that the human-interpretability of linked data does not need to be sacrificed—it can actually be enhanced—by considering “charts as metadata.” In addition, we show how knowledge graphs and interactive data visualization, when combined, offer a mutually beneficial way to explore and share scientific knowledge.
Read the full, open-access article in Scientific Data at:
Deagen, M. E., McCusker, J. P., Fateye, T., Stouffer, S., Brinson, L. C., McGuinness, D. L., & Schadler, L. S. FAIR and Interactive Data Graphics from a Scientific Knowledge Graph. Sci. Data. 9, 239, https://doi.org/10.1038/s41597-022-01352-z (2022).






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in